The first operation is to read in the response matrix. To do this one has to define a model and to "fakeit none":
XSPEC> mo pow
Input parameter value, delta, min, bot, top, and max values for ...
Mod parameter 1 of component 1 powerlaw PhoIndex2 variable fit parameters
1.000 1.0000E-02 -3.000 -2.000 9.000 10.00
Mod parameter 2 of component 1 powerlaw norm
1.000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
---------------------------------------------------------------------------
mo = powerlaw[1]
Model Fit Model Component Parameter Value
par par comp
1 1 1 powerlaw PhoIndex 1.00000 +/- 0.
2 2 1 powerlaw norm 1.00000 +/- 0.
---------------------------------------------------------------------------
XSPEC>fakeit none
For fake data, file # 1 needs response file: pds.rmf
Use counting statistics in creating fake data? (y)
Input optional fake file prefix (max 12 chars):
Override default values for file # 1
Fake data filename (pds.fak):
T, A, Bkg, cornorm ( 1.0000 , 1.0000 , 1.0000 , 0. ): 10000
Net count rate (cts/cm^2/s) for file 1 1.922 +/- 1.3864E-02
Chi-Squared = 17.68 using 24 PHA bins.
Reduced chi-squared = 0.8036
Now read in the effective area and background files:
XSPEC> arf pds.arf
Chi-Squared = 8.6504E+09 using 24 PHA bins.
Reduced chi-squared = 3.9320E+08
XSPEC> back bkg_pds_nostat.pha
Net count rate (cts/cm^2/s) for file 1 -30.89 +/- 1.7985E-02(*****% total)
Chi-Squared = 6.1414E+09 using 24 PHA bins.
Reduced chi-squared = 2.7916E+08
Now put to zero the model normalization and "fakeit" to apply background statistics.
** NOTE ** If you are using a ULTRIX machine make sure to set the normalization to a very small value (e.g. 1.e-7) but NOT to 0. This is to avoid underflow errors.
XSPEC> newpar 2 0
2 variable fit parameters
Chi-Squared = 4.1434E+06 using 24 PHA bins.
Reduced chi-squared = 1.8834E+05
XSPEC> fakeit
Use counting statistics in creating fake data? (y) y
Input optional fake file prefix (max 12 chars):
Override default values for file # 1
Fake data filename (pds.fak):
T, A, Bkg, cornorm ( 10000. , 1.0000 , 1.0000 , 0. ): 50000
Net count rate (cts/cm^2/s) for file 1 -1.7398E-02+/- 2.1427E-02( -0.1% total)
Chi-Squared = 20.66 using 24 PHA bins.
Reduced chi-squared = 0.9392
The file pds.fak now contains the background for a 50,000 PDS observation with
the right poisson statistics.
XSPEC> mo wa cutoffpl
Chose your favourite models, set the right model normalization and
"fakeit" again:
Input parameter value, delta, min, bot, top, and max values for ...
4 variable fit parameters
Mod parameter 1 of component 1 wabs nH 10^22
1.000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
1
Mod parameter 2 of component 2 cutoffpl PhoIndex
1.000 1.0000E-02 -3.000 -2.000 9.000 10.00
1.8
Mod parameter 3 of component 2 cutoffpl HighECut
15.00 1.0000E-02 1.0000E-02 1.000 50.00 200.0
50
Mod parameter 4 of component 2 cutoffpl norm
1.000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
0.1
---------------------------------------------------------------------------
mo = wabs[1] (cutoffpl[2])
Model Fit Model Component Parameter Value
par par comp
1 1 1 wabs nH 10^22 1.00000 +/- 0.
2 2 2 cutoffpl PhoIndex 1.80000 +/- 0.
3 3 2 cutoffpl HighECut 50.0000 +/- 0.
4 4 2 cutoffpl norm 0.100000 +/- 0.
---------------------------------------------------------------------------
Chi-Squared = 5.6645E+04 using 24 PHA bins.
Reduced chi-squared = 2832.
XSPEC> fakeit
Use counting statistics in creating fake data? (y) y
Input optional fake file prefix (max 12 chars):
Override default values for file # 1
Fake data filename (pds.fak):
T, A, Bkg, cornorm ( 50000. , 1.0000 , 1.0000 , 0. ):
pds.fak exists, reuse it ? y
Net count rate (cts/cm^2/s) for file 1 3.409 +/- 2.9250E-02( 9.4% total)
Chi-Squared = 15.75 using 24 PHA bins.
Reduced chi-squared = 0.7875
Now you should have your simulated spectrum in the file pds.fak. The parameters used in this simulation are appropriate for NGC4151 in a high state. You can look at the count rates predicted and simulated with "show" and look at the spectrum with "setplot energy" and then "plot data". You can simulate the spectra in the other SAX instruments following the same procedure.
Then, you may want to try to fit the simulated spectra
(MECS 10 ks exposure + PDS 50 ks exposure in this example) with a
simple power law, to understand to which extent the cutoff is detectable in
a source like NGC4151.
Read the spectra in XSPEC:
XSPEC> data 1:1 mecs.fak
*WARNING*: Background and data file Detector ID mismatch
(SAX MECS(*3),SAX ME SAX ME) for file 1
*WARNING*: Response and data file detector ID mismatch
(SAX ME,SAX ME SAX ME) for file 1
*WARNING*: Aux. Resp. and data file detector ID mismatch
(SAX ME,SAX ME SAX ME) for file 1
Net count rate (cts/cm^2/s) for file 1 4.601 +/- 2.1462E-02( 99.9% tot)
XSPEC> data 2:2 pds.fak
*WARNING*: Background and data file Detector ID mismatch
(SAX PDS,SAX PDS SAX PDS) for file 2
*WARNING*: Response and data file detector ID mismatch
(SAX PDS,SAX PDS SAX PDS) for file 2
*WARNING*: Aux. Resp. and data file detector ID mismatch
(SAX PDS,SAX PDS SAX PDS) for file 2
Net count rate (cts/cm^2/s) for file 2 3.409 +/- 2.9250E-02( 9.4% tot)
XSPEC> back 2:2 bgd_pds_50.pha
*WARNING*: Background and data file Detector ID mismatch
(SAX PDS,SAX PDS SAX PDS) for file 2
Net count rate (cts/cm^2/s) for file 2 3.393 +/- 5.7870E-02( 9.4% tot)
XSPEC> mo wa pow
Input parameter value, delta, min, bot, top, and max values for ...
Mod parameter 1 of component 1 wabs nH 1 for data group 1
1.000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
Mod parameter 2 of component 2 powerlaw PhoI for data group 1
1.000 1.0000E-02 -3.000 -2.000 9.000 10.00
Mod parameter 3 of component 2 powerlaw norm for data group 1
1.000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
0.1
Mod parameter 4 of component 3 wabs nH 1 for data group 2
1.000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
Mod parameter 5 of component 4 powerlaw PhoI for data group 2
1.000 1.0000E-02 -3.000 -2.000 9.000 10.00
Mod parameter 6 of component 4 powerlaw norm for data group 2
0.1000 1.0000E-03 0. 0. 1.0000E+05 1.0000E+06
0.2
---------------------------------------------------------------------------
mo = wabs[1] (powerlaw[2])
Model Fit Model Component Parameter Value Data
par par comp group
1 1 1 wabs nH 10^22 1.00000 +/- 0. 1
2 2 2 powerlaw PhoIndex 2.00000 +/- 0. 1
3 3 2 powerlaw norm 0.100000 +/- 0. 1
4 1 3 wabs nH 10^22 2.00000 = par 1 2
5 2 4 powerlaw PhoIndex 1.00000 = par 2 2
6 4 4 powerlaw norm 0.200000 +/- 0. 2
---------------------------------------------------------------------------
4 variable fit parameters
Chi-Squared = 3.2685E+07 using 164 PHA bins.
Reduced chi-squared = 2.0428E+05
XSPEC> fit
Chi-Squared Lvl Fit param # 1 2 3 4
839.12 -3 1.528 2.144 0.1417 0.1889
411.71 -4 1.458 2.127 0.1509 0.1673
410.02 -5 1.426 2.109 0.1462 0.1586
409.99 -6 1.429 2.110 0.1467 0.1595
409.99 -7 1.428 2.110 0.1466 0.1595
---------------------------------------------------------------------------
mo = wabs[1] (powerlaw[2])
Model Fit Model Component Parameter Value Data
par par comp group
1 1 1 wabs nH 10^22 1.42840 +/- 0.36275E-01 1
2 2 2 powerlaw PhoIndex 2.11035 +/- 0.15243E-01 1
3 3 2 powerlaw norm 0.146644 +/- 0.36999E-02 1
4 1 3 wabs nH 10^22 1.42840 = par 1 2
5 2 4 powerlaw PhoIndex 2.11035 = par 2 2
6 4 4 powerlaw norm 0.159489 +/- 0.76906E-02 2
---------------------------------------------------------------------------
Chi-Squared = 410.0 using 164 PHA bins.
Reduced chi-squared = 2.562
XSPEC> cpd /xterm
XSPEC> plot data delchi
XSPEC> setplot energy
XSPEC> plot data delchi
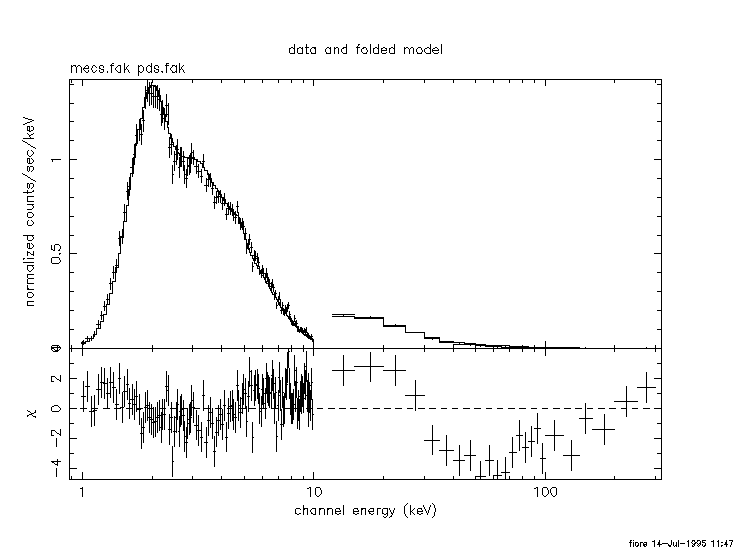
The fit is clearly unacceptable, as evident from the following plot:
Please send questions/problem reports to: helpdesk@sax.sdc.asi.it
Last Revised: July 14, 1995